AUCTORES
Globalize your Research
Research Article | DOI: https://doi.org/10.31579/2766-2314/130
Protein Research Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), Alexandria, Egypt.
*Corresponding Author: Nawal Abd El-Baky, Protein Research Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), Alexandria, Egypt.
Citation: Nawal Abd El-Baky, Amro A. Amara, (2024), Comparative Investigation of some Bioeffective Short Peptides of Marine Derivation, J, Biotechnology and Bioprocessing, 5(2); DOI:10.31579/2766-2314/130
Copyright: © 2024, Nawal Abd El-Baky. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 22 January 2024 | Accepted: 31 January 2024 | Published: 08 February 2024
Keywords: cyclic peptides; hybrids or consensus; intrinsic disorder; marine bioeffective peptides; physicochemical properties
Short peptides especially marine ones are unique in their amino acids constituents that build their structure/function/specificity. Their physical and chemical properties make them diverse in their bioeffectiveness. In this study, different bioeffective peptides from marine bio-resources were investigated to map distinctive properties in their structure that could affect their function and specificity. The investigated short peptides were Pardaxins (1-5), Arenicin (1-2), Clavanin-(A-E), Aurelin, Dicynthaurin, Halocidin subunit (A-B), Tachyplesin (II-III), and Polyphemusin (I-II). Their different sequences were analyzed comparatively through alignment, their clustering (phylogeny tree), amino acids-%, and some other physical and chemical parameters. For each of them, cyclic structure based on disulfide bonds if present, and intrinsic disorder, were inspected. First alignment revealed significant diversion among their sequences, while removing unshaded amino acids from the alignment made them closer in their phylogenetic relationships and more evident to design hybrids or consensus of sequences and functions. Polyphemusin I and II had the highest charges (net) with values of +7, yet Pardaxins 1, 3, and 5 had 0 charges. All of them had higher numbers of hydrophilic amino acids than hydrophobic ones except for Aurelin, Tachyplesin II and III, and Polyphemusin I and II. Seven from twenty studied peptides had cyclic structures via disulfide bond between their cysteine residues. One amino acid was not found in all sequences; methionine. All sequences displayed other missed amino acids than methionine. Dicynthaurin is a naturally ordered peptide (via PONDR score), but Halocidin subunit B was totally disordered. This study paves the way for designing hybrids or consensus of these peptides in sequences and functions depending on their distinctive features.
Marine organisms are a vast cradle of unusual bioeffective peptides. Peptides from several aquatic species with extraordinary impending nutraceutical and medicinal values are the emphasis of up-to-date research. Their crucial biological roles including microbicidal or antioxidative action, antitumor efficiency, cardioprotective properties, immunomodulatory role, etc. lead to continuous efforts and attempts to design and formulate these peptides (Ngo et al., 2012, Cheung et al., 2015, Ovchinnikova, 2019). Marine bioeffective peptides are distinguished from non-marine-derived ones by their resistance to proteolysis performed by proteases of gastrointestinal tract due to steric factors caused by their cyclic nature (Pangestuti and Kim, 2017, Saint Jean et al., 2017, Ibrahim et al., 2018, Phyo et al., 2018).
Marine bioeffective peptides are diverse and have different characteristics. Pardaxin possess antimicrobial actions and comprises 33 amino acids (Kolusheva et al., 2008). It is also utilized for repelling sharks (Shai, 1994). Pardaxins 1, 2 and 3 were produced from Pardachirus pavoninus, and Pardaxins 4 and 5 were released by Pardachirus marmoratus (Shai et al., 1988). Pardaxin resembles melittin in its ability to lyse cells of bacteria and mammals (Oren and Shai, 1996). It might prompt apoptosis, leading to antitumor efficacy (Hsu et al., 2011). Arenicin is bactericidal to Gram-negative bacteria and induced apoptosis (Cho and Lee, 2011, Krenev et al., 2020). It also showed antifungal effects (Park et al., 2011). Arenicin 1 and 2 were isolated from Arenicola marina (Ovchinnikova et al., 2004, Umnyakova et al., 2018).
Aurelin was isolated from Aurelia aurita (jellyfish) and exhibited antimicrobial functions (Ovchinnikova et al., 2006, Shenkarev et al., 2012). Aurelin shared similar features in its structure with defensins and toxins that block channels (Ovchinnikova et al., 2006). It displayed bactericidal effectiveness on gram-negative and -positive bacteria (Ovchinnikova et al., 2006).
Clavanins (Clavanin-A, -B, -C, -D and –E) were obtained from Styela clava (Tunicate) hemocytes and revealed antimicrobial efficiency (Juliano et al., 2017, Lehrer et al., 2001, Menzel et al., 2002). Their antimicrobial functions, which rely on their α-helical structures, were thoroughly investigated against various microbes and even viruses such as herpes simplex virus 1 (HSV-1) (Lee et al., 1997, Lehrer et al., 2001, Carriel-Gomes et al., 2007). Clavanin-A antimicrobial efficiency contributes to innate immune responses of Styela clava (Juliano et al., 2017). Besides, it caused in vitro viral inactivation of non-enveloped adenovirus and rotavirus (Carriel-Gomes et al., 2007). Clavanin-B was proved to have antiviral effectiveness on HIV (Ireland et al., 2008).
Dicynthaurin, another marine bioeffective peptide possessing antimicrobial activity, has been isolated from Halocynthia aurantium (Lee et al., 2001a). It demonstrated expansive action on Gram +ve and Gram -ve bacteria nonetheless was not active against Candida albicans (Lee et al., 2001a). Halocidin was also obtained from Halocynthia aurantium and has bactericidal and anti-Candida actions (Jang et al., 2003, Jang et al., 2006, Jang et al., 2002). A peptide derived from Halocidin could effectively treat oral infection with Candida albicans in mice (Shin et al., 2013).
Polyphemusins from Limulus polyphemus have antimicrobial efficacy (Marggraf et al., 2018, Lee et al., 2022). They are potent against various microbes and viruses (Lee et al., 2022, Panteleev et al., 2020, Tziveleka et al., 2003). Tachyplesins antimicrobial peptides were released from hemocytes of horseshoe crab (Ohta et al., 1992).Tachyplesin I could penetrate cells and has anticancer properties (Vernen et al., 2019a).
In the present investigation, twenty bioeffective peptides (Pardaxins (1-5), Arenicin (1-2), Clavanin-(A-E), Aurelin, Dicynthaurin, Halocidin subunit (A-B), Tachyplesin (II-III), and Polyphemusin (I-II)) from marine bio-resources were explored to record distinctive properties in their structures affecting their antimicrobial functions. Their different sequences were compared to each other, in addition to amino acids-%, and some other chemical and physical parameters. For each of them, intrinsic disorder and cyclic structures based on disulfide bonds were also inspected. The obtained data can lead to designing of hybrids or consensus of these peptides that may achieve more potent actions or even new functions.
Investigated marine bioeffective peptides
The twenty marine bioeffective peptides included in the present work are found in Table 1.
Pardaxins (1-5), 1, Arenicin 2, Clavanin-A, Clavanin-B, Clavanin-E, Clavanin-C, Clavanin-D, Aurelin, Dicynthaurin, Halocidin subunit A, Halocidin subunit B, Tachyplesin-II, Polyphemusin I, Tachyplesin III, Polyphemusin II have lengths of 33, 21, 21, 23, 23, 22, 23, 22, 40, 30, 18, 15, 17, 18, 16, and 18 amino acids, respectively.
| Peptide parent name | Specific name | Sequence |
| Pardaxins | Pardaxins l | GFFALIPKIISSPLFKTLLSAVGSALSSSGEQE |
| Pardaxins 2 | GFFALIPKIISSPIFKTLLSAVGSALSSSGGQE | |
| Pardaxins 3 | GFFAFIPKIISSPLFKTLLSAVGSALSSSGEQE | |
| Pardaxins 4 | GFFALIPKIISSPLFKTLLSAVGSALSSSGGQE | |
| Pardaxins 5 | GFFALIPKIISSPLFKTLLSAVGSALSSSGDQE | |
| Arenicin | Arenicin 1 | RWCVYAYVRVRGVLVRYRRCW |
| Arenicin 2 | RWCVYAYVRIRGVLVRYRRCW | |
| Clavanins | Clavanin-A | VFQFLGKIIHHVGNFVHGFSHVF |
| Clavanin-B | VFQFLGRIIHHVGNFVHGFSHVF | |
| Clavanin-C | VFHLLGKIIHHVGNFVYGFSHVF | |
| Clavanin-D | FKLLGRIIHHVGNFVYGFSHVF | |
| Clavanin-E | FKLLGKIIHHVGNFVHGFSHVF | |
| Aurelin | Aurelin | AACSDRAHGHICESFKSFCKDSGRNGVKLRANCKKTCGLC |
| Dicynthaurin | Dicynthaurin | ILQKAVLDCLKAAGSSLSKAAITAIYNKIT |
| Halocidin | Halocidin subunit A | WLNALLHHGLNCAKGVLA |
| Halocidin subunit B | ALLHHGLNCAKGVLA | |
| Tachyplesin | Tachyplesin II | RWCFRVCYRGICYRKCR |
| Tachyplesin III | WCFRVCYRGICYRKCR | |
| Polyphemusin | Polyphemusin I | RRWCFRVCYRGFCYRKCR |
| Polyphemusin II | RRWCFRVCYKGFCYRKCR |
Table 1: The studied peptides sequences.
Peptide sequences comparison with Ugene
Alignment No. 1
The up-to-date free version of Ugene 49.1 (Portable) was used to do the peptides sequences comparison (alignment) (Okonechnikov et al., 2012, Rose et al., 2018). This alignment was carried out by the alignment selection with Clustal Omega in the software since the investigated peptides had different lengths. The shading was done for amino acids with analogous physicochemical features.
Alignment No. 2
Aiming to increase the probability of finding close associations between the twenty peptides sequences to facilitate planning of peptides hybrids, the unshaded amino acids after using the physicochemical shading selection were eliminated. The obtained sequences were aligned once more and even adjusted manually afterwards. This created reduced hypothetical peptides sequences.
After running each of the fore-mentioned alignments, the phylogeny tree was generated. The involved mode in tree building was Neighbor Joining and distance matrix was Jones-Taylor-Thornton.
Recording the properties of the twenty peptides’ complete sequences
The peptides general features, which included mass (Da), isoelectric point, net charge, hydrophobicity, and extinction coefficients were attained from the PepDraw web site (https://pepdraw.com).
Drawing cyclic peptides structures based on disulfide bonds
If studied peptides had cyclic structures via disulfide bond between cysteine residues was detected via PepSMI (https://www.novoprolabs.com/tools/convert-peptide-to-smiles-string).
Concluding the amino acids-% in the investigated peptides
DAMBE 7.2.102 was utilized to estimate the amino acids-% in each of the twenty peptides sequences (complete ones not reduced hypothetical peptides sequences) under investigation (Xia, 2018).
Data plotting of amino acids-%
The data of amino acids-% were considered in PAST 4.14. software using the clustering analysis option (Hammer et al., 2001). Neighbor joining was chosen with Hoen similarity index to cluster comparable peptide groups.
The software plotted these data to elucidate the variations within amino acids or within the peptides. It also enabled contrasts between the diverse amino acids as well as viewing the standard deviations of their existence % among peptides, which mirrored the distinction within different peptides constituent from each amino acid.
Helical Wheel (HW)
Anthreprot 6.9.3. determined the HW of each peptide sequence to record the hydrophobic amino acids and hydrophilic ones on the surface of HW and their distribution through the complete peptides sequences and reduced hypothetical ones (Deléage et al., 1989, Deléage et al., 1988, Deléage et al., 2001, Deléage, 2012).
Specifying the amino acids that had disorder
All sequences were loaded in the PrDOS server to find any amino acid that has disorder or form with other ones, a disorder region. PrDOS accepted all the sequences lengths involved in this study (Ishida and Kinoshita, 2007). In addition, PONDR (www.pondr.com) was used to find natural disorder compared to crystallization data of natural proteins but in peptides that have sequence length of ≥ 30 amino acids. flDPnn was used to map propensity of disorder for investigated peptides (Hu et al., 2021).
Peptides and specifically peptides from marine derivation showed unlimited potentials (Rawat et al., 2006, Liu et al., 2015, Simonetti et al., 2009, De Clercq, 2000). They found their way in different applications including fungicidal (Jang et al., 2006, Fan et al., 2023, Han et al., 2016), bactericial (Ohta et al., 1992, Panteleev et al., 2017, Saito et al., 1995, Serna et al., 2021, Li et al., 2020), virucidal (even to SARS-CoV-2) (Gogineni et al., 2015, Xie et al., 2016, Riccio et al., 2020, Raihan et al., 2021), anticancer (Vernen et al., 2019a, Ahmed et al., 2021, Vernen et al., 2019b, Zhang et al., 2021, Han et al., 2015, Lin et al., 2013), etc.
This study involved random selection of twenty bioeffective peptides from marine derivation; Arenicin (Arenicin 1 and 2) (Cho and Lee, 2011, Krenev et al., 2020, Park et al., 2011, Ovchinnikova et al., 2004, Umnyakova et al., 2018), Aurelin (Ovchinnikova et al., 2006, Shenkarev et al., 2012), Clavanins (-A, -B, -C, -D and -E) (Silva et al., 2015), Dicynthaurin (Bringezu et al., 2007b, Lee et al., 2001b, Bringezu et al., 2007a, Majerowicz et al., 2007, Wen et al., 2007), Halocidin subunit A and subunit B (Shin et al., 2010, Han et al., 2016), Pardaxins 1-5 (Oren and Shai, 1996), Polyphemusin I and II (Marggraf et al., 2018, Lee et al., 2022), and Tachyplesin II and III (Ohta et al., 1992, Vernen et al., 2019a). Their sequences were compared to each other, in addition to amino acids-%, and other chemical and physical parameters. Their disorder properties and cyclic structures based on disulfide bonds were also checked.
Alignment No. 1
Firstly, we performed the classical step to compare their sequences; alignment. Apparently, the peptides sequences were confusing to the alignment softwares because of their different lengths, and none of them gave the same result (data not shown). Meanwhile, Clustal-Omega gave more appropriate results (Figure 1), where similarity could be observed, even if the amino acids was still scattered. This gave us the idea to use the physicochemical properties mode to shade the similar amino acids and to eliminate the unshaded ones in the following run.
Figure 1 revealed that peptides from the same source were aligned together. For example, Pardaxins 1 to 5 have been all aligned as a separate cluster. Besides, Tachyphemusin II and III were aligned with Polyphemusin I and II along with Arenicin 1 and 2. In the middle, Clavanin A-E, Aurelin, Halocidin subunit A and B showed similarity. In fact, Clustal Omega exhibited perfect grouping for analogous regions.
Furthermore, the shadding was based on the physiochemical properties of the amino acids (Figure 1).
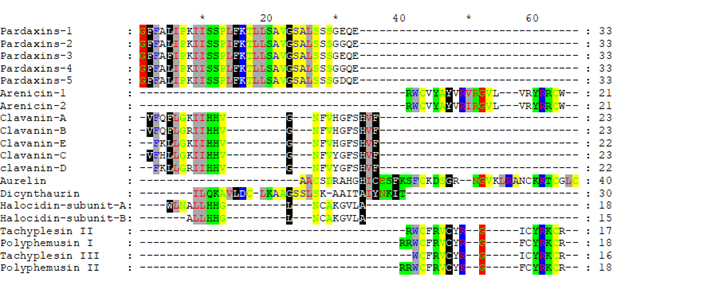
Figure 1: Multiple alignment of the complete sequences of the investigated peptides with Clustal Omega (Ugene software).
Amino acids that appeared without coloring (either did not have unique physiochemical characteristics at their position or did not align with a region of distinctive physciochemical features) were removed and the reduced hypothetical sequences were realigned.
Alignment No. 2
After removing the unshaded amino acids and performing a second alignment run, the similarity became more obvious (Figure 2 and Figure 3). If Arenicin 1 and 2 was moved down manually, all of the hypothetical sequences would align together in different regions as in Figure 3. Such approach might be useful in designing peptides de novo or hybrids.
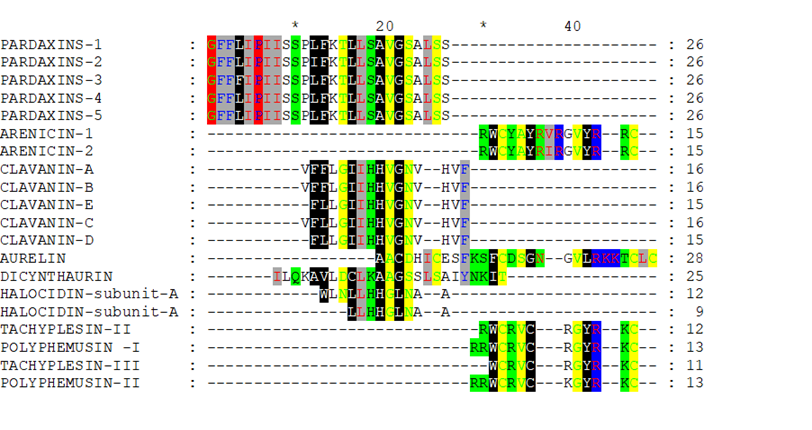
Figure 2: Multiple alignment of the reduced hypothetical sequences of the investigated peptides.
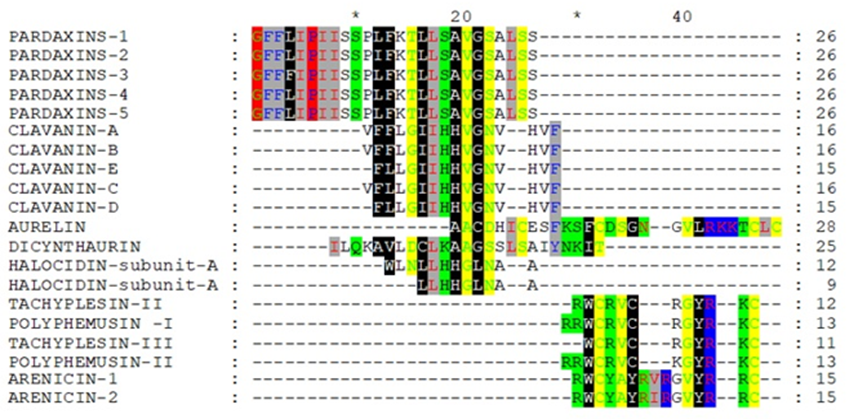
Figure 3: Multiple alignment of the reduced hypothetical sequences of the investigated peptides after adjustment manually.The phylogeny tree obtained from Ugene of the aligned twenty peptides sequences confirmed that removing unshaded amino acids increased the clustering of peptides of different classes together (Figure 4, and Figure 5). The cadre showed grouping of the peptides (Arenicin 2, Halocidin subunit A and B, Clavanin-B, Tachyplesin III, Clavanin-E, Polyphemusin I, Clavanin-A, Tachyplesin II, Clavanin-C, Polyphemusin II, and Clavanin- D) due to similarity after reducing the original sequence (Figure 5).

Figure 4: Phylogeny tree from the alignment of complete peptides sequences.

Figure 5: Phylogeny tree from the alignment of reduced hypothetical peptides sequences. The cadre showed grouping of the peptides due to similarity after reducing the original sequence.
Recording the properties of the twenty peptides’ complete sequences
Table 2 illustrated the properties of the twenty peptides amino acids sequences. Aurelin had the highest mass of 4300 Da and uppermost hydrophobicity of +45.67 Kcal * mol-1 among all peptides (Table 2). Polyphemusin I and II had the utmost charges (net) with values of +7, yet Pardaxins 1, 3, and 5 had 0 charges (Table 2). Arenicin 1 and 2 exhibited the leading isoelectric point of 11.14 (Table 2).
| Peptide Name | Length | Mass (Da) | isoelectric point (pI) | Net charge | Hydrophobicity Kcal * mol-1 | Extinction coefficient M-1* cm-I | Extinction coefficient2 M-1* cm-I |
| Arenicin 1 | 21 | 2758.46 | 11.14 | +6 | +10.51 | 15470 | 15595 |
| Arenicin 2 | 21 | 2772.48 | 11.14 | +6 | +9.85 | 15595 | 15470 |
| Aurelin | 40 | 4300.00 | 8.94 | +5 | +45.67 | 375 | 0 |
| Clavanin-A | 23 | 2665.40 | 9.93 | +1 | +11.67 | 0 | 0 |
| Clavanin-B | 23 | 2693.40 | 10.90 | +1 | +10.6 | 0 | 0 |
| Clavanin-C | 23 | 2666.42 | 9.59 | +1 | +10.65 | 1490 | 1490 |
| Clavanin-D | 22 | 2586.40 | 10.38 | +2 | +10.59 | 1490 | 1490 |
| Clavanin-E | 22 | 2532.39 | 10.59 | +2 | +14.62 | 0 | 0 |
| Dicynthaurin | 30 | 3103.76 | 9.91 | +3 | +19.72 | 1490 | 1490 |
| Halocidin subunit A | 18 | 1929.04 | 9.04 | +1 | +12.04 | 5500 | 5500 |
| Halocidin subunit B | 15 | 1515.83 | 9.04 | +1 | +14.53 | 0 | 0 |
| Pardaxins 1 | 33 | 3393.84 | 6.97 | 0 | +15.03 | 0 | 0 |
| Pardaxins 2 | 33 | 3321.82 | 9.95 | +1 | +12.68 | 0 | 0 |
| Pardaxins 3 | 33 | 3427.82 | 6.97 | 0 | +14.57 | 0 | 0 |
| Pardaxins 4 | 33 | 3321.82 | 9.95 | +1 | +12.55 | 0 | 0 |
| Pardaxins 5 | 33 | 3379.82 | 6.93 | 0 | +15.04 | 0 | 0 |
| Polyphemusin I | 18 | 2457.18 | 10.54 | +7 | +15.24 | 8730 | 8480 |
| Polyphemusin II | 18 | 2429.17 | 10.33 | +7 | +16.23 | 8730 | 8480 |
| Tachyplesin II | 17 | 2267.09 | 10.01 | +6 | +14.02 | 8730 | 8480 |
| Tachyplesin III | 16 | 2110.99 | 9.48 | +5 | +12.21 | 8730 | 8480 |
Table 2: Properties of the different peptides’ amino acids sequences.
Drawing cyclic peptides structures based on disulfide bonds
Seven studied peptides (Arenicin 1 and 2, Polyphemusin I and II, Tachyplesin II and III, and Aurelin) had cyclic structures via disulfide bond between cysteine residues (Figure 6). The other thirteen sequences formed only linear structures due to the absence of cysteine residues (Pardaxins l-5 and Clavanin-A-E) or presence of only one cysteine (Dicynthaurin, and
Halocidin subunit A and B). The most unique cyclic structure was for Aurelin as the peptide contains 6 cysteine residues (Figure 6). These cyclic structures could make them more unaffected by proteolysis performed by proteases of gastrointestinal tract due to steric factors (Pangestuti and Kim, 2017, Saint Jean et al., 2017, Ibrahim et al., 2018, Phyo et al., 2018).
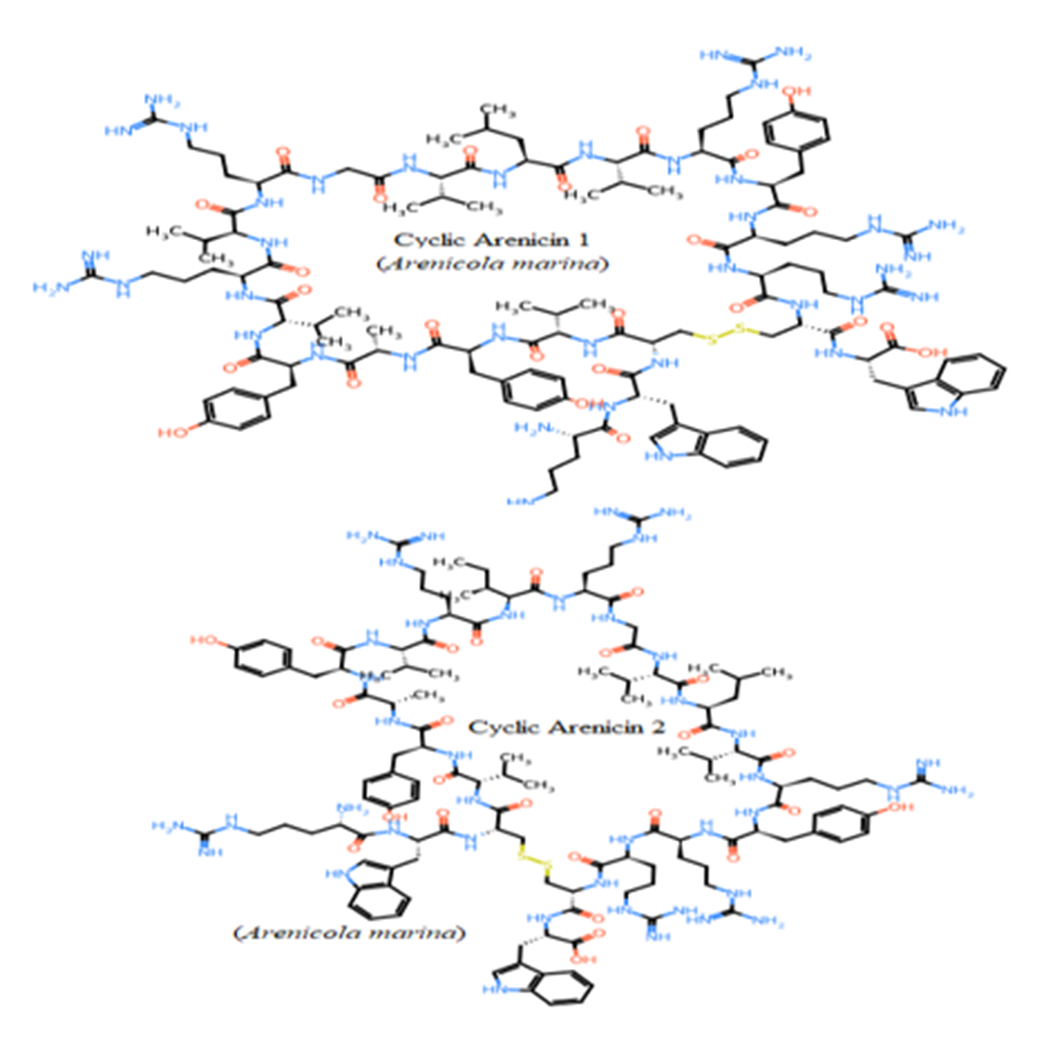

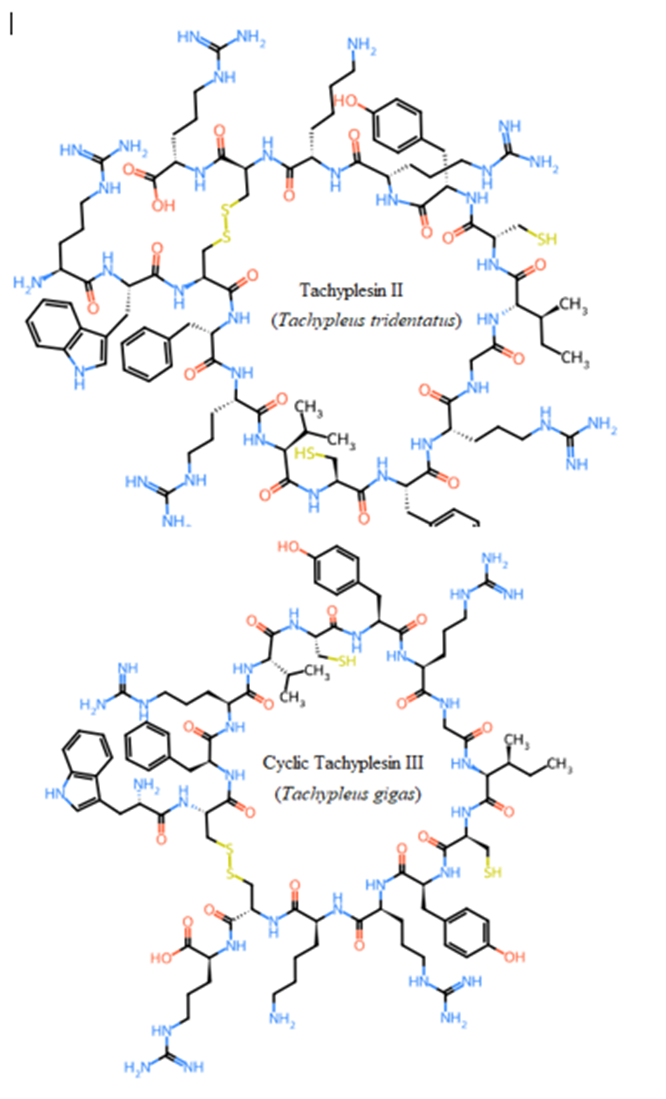
Figure 6: Unique cyclic structures of seven from twenty studied peptides via disulphide bond between cysteines
Concluding the amino acids-% in the investigated peptides
To deepen our understanding to the 20 selected marine bioeffective peptides and their amino acids constituents, amino acid-% distribution was checked.
Stacked chart of the amino acids-% of investigated peptides in Figure 7 displayed that methionine did not exist at all in all of the 20 peptides.
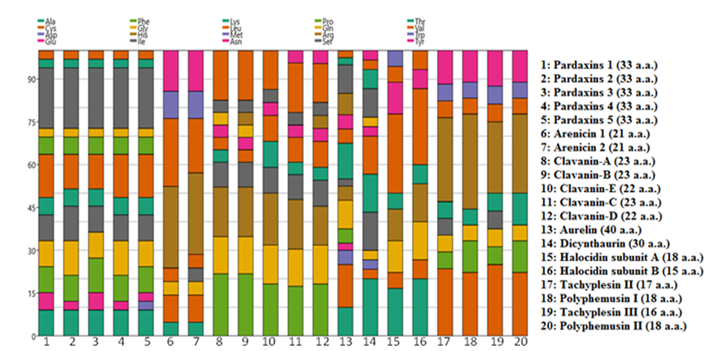
Figure 7: Stacked chart of the amino acids-% of investigated peptides.
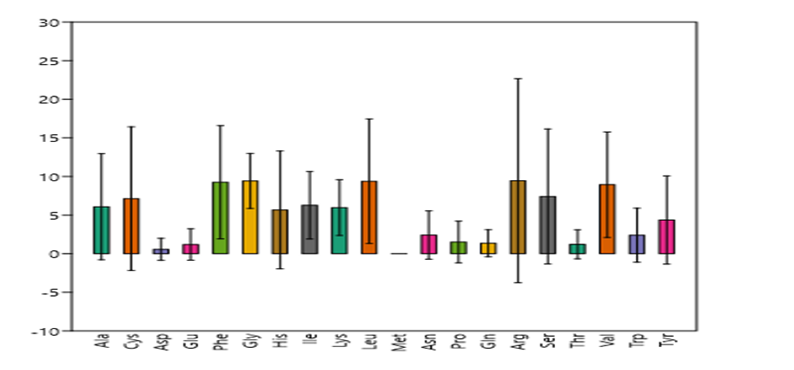
Figure 8: Amino acids-% in investigated peptides with standard deviation
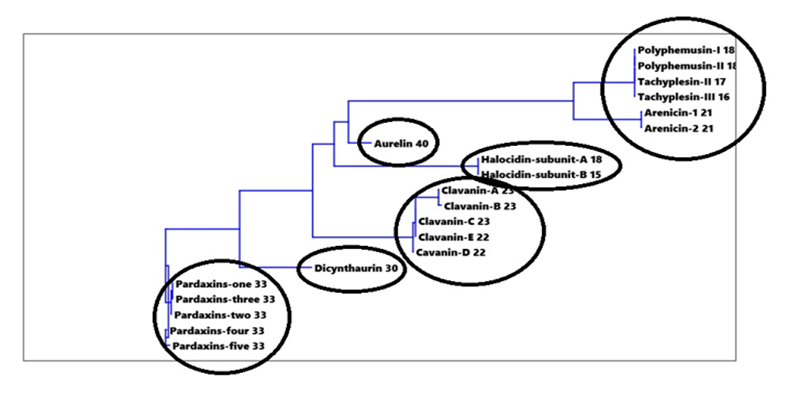
Figure 9: Cluster analysis for amino acids-% in investigated peptides
Demonstrated amino acids-% in investigated peptides with standard deviation of their existence % among peptides, which mirrored the distinction within different peptides constituent from each amino acid. This brief statistical estimation revealed the discrepancy among the amino acids as well as within each peptide class.
The composition of amino acids is strictly associated with bioeffectiveness of these bioeffective peptides. For instance, Arenicin 1 was found to have fungicidal efficacy that is dependent on tryptophan in its N terminus and arginine in its β-turn (Park et al., 2011).
Cluster analysis for amino acids-% in investigated peptides in Figure 9 indicated that the twenty amino acids-% classified peptides to six major groups, where Pardaxins 1-5 were clustered in one group. The same case was found with Clavanin-A, -B, -C, -D and -E. Dicynthaurin and Aurelin each could be considered as separate groups (Figure 9). Polyphemusin I and II, Tachyplesin II and III along with Arenicin 1 and 2 are grouped in one cluster. While, Halocidin subunit A and B were clustered together as in Figure 9.
Helical Wheel (HW)
The HW analysis investigated the distribution of the hydrophobic and hydrophilic amino acids on helical folding form of peptides. The results were summarized in Table 3. Table 3 showed that all peptides had higher numbers of hydrophilic amino acids than hydrophobic ones except for Aurelin, Tachyplesin II and III, and Polyphemusin I and II. Hydrophobic amino acids in peptides have critical contribution to their bactericidal effectiveness as they enhance peptides ability to bind and disintegrate membranes of bacteria (Saint Jean et al., 2017). The overall differences between the numbers of the hydrophobic and hydrophilic amino acids ranged from two to four (Table 3). Meanwhile, the oddest difference was in case of Aurelin, which showed differences equal to 10 amino acids (Table 3)
NAME | HYDROPHOBIC AND HYDROPHILIC AMINO ACIDS DISTRIBUTION | NO. OF HYDROPHOBIC AMINO ACIDS | NO. OF HYDROPHILIC AMINO ACIDS |
| PARDAXINS 1 | GFFALIPKIISSPLFKTLLSAVGSALSSSGEQE | 13 | 15 |
| PARDAXINS 2 | GFFALIPKIISSPIFKTLLSAVGSALSSSGGQE | 12 | 15 |
| PARDAXINS 3 | GFFAFIPKIISSPLFKTLLSAVGSALSSSGEQE | 13 | 15 |
| PARDAXINS 4 | GFFALIPKIISSPLFKTLLSAVGSALSSSGGQE | 12 | 15 |
| PARDAXINS 5 | GFFALIPKIISSPLFKTLLSAVGSALSSSGDQE | 13 | 15 |
| ARENICIN 1 | RWCVYAYVRVRGVLVRYRRCW | 6 | 9 |
| ARENICIN 2 | RWCVYAYVRIRGVLVRYRRCW | 6 | 9 |
| CLAVANIN-A | VFQFLGKIIHHVGNFVHGFSHVF | 8 | 11 |
| CLAVANIN-B | VFQFLGRIIHHVGNFVHGFSHVF | 8 | 11 |
| CLAVANIN-E | FKLLGKIIHHVGNFVHGFSHVF | 8 | 11 |
| CLAVANIN-C | VFHLLGKIIHHVGNFVYGFSHVF | 7 | 11 |
| CLAVANIN-D | FKLLGRIIHHVGNFVYGFSHVF | 7 | 11 |
| AURELIN | AACSDRAHGHICESFKSFCKDSGRNGVKLRANCKKTCGLC | 20 | 10 |
| DICYNTHAURIN | ILQKAVLDCLKAAGSSLSKAAITAIYNKIT | 12 | 15 |
| HALOCIDIN SUBUNIT A | WLNALLHHGLNCAKGVLA | 5 | 10 |
| HALOCIDIN SUBUNIT B | ALLHHGLNCAKGVLA | 4 | 8 |
| TACHYPLESIN II | RWCFRVCYRGICYRKCR | 6 | 4 |
| POLYPHEMUSIN I | RRWCFRVCYRGFCYRKCR | 7 | 4 |
| TACHYPLESIN III | WCFRVCYRGICYRKCR | 5 | 4 |
| POLYPHEMUSIN II | RRWCFRVCYKGFCYRKCR | 7 | 4 |
Table 3:The hydrophobic amino acids (red) and hydrophilic ones (blue) distribution through the complete peptides sequences.
Table 4 demonstrated the hydrophobic amino acids (red) and hydrophilic ones (blue) distribution through reduced hypothetical peptides sequences. The numbers of hydrophobic amino acids was significantly reduced after sequence reductions.
Name | Hydrophobic and hydrophilic amino acids distribution | No. of hydrophobic amino acids | No. of hydrophilic amino acids |
| Pardaxins 1 | GFFLIPIISSPLFKTLLSAVGSALSS | 8 | 14 |
| Pardaxins 2 | GFFLIPIISSPIFKTLLSAVGSALSS | 8 | 14 |
| Pardaxins 3 | GFFFIPIISSPLFKTLLSAVGSALSS | 8 | 14 |
| Pardaxins 4 | GFFLIPIISSPLFKTLLSAVGSALSS | 8 | 14 |
| Pardaxins 5 | GFFLIPIISSPLFKTLLSAVGSALSS | 8 | 14 |
| Arenicin 1 | RWCYAYRVRGVYRRC | 5 | 4 |
| Arenicin 2 | RWCYAYRIRGVYRRC | 5 | 4 |
| Clavanin-A | VFFLGIIHHVGNVHVF | 4 | 8 |
| Clavanin-B | VFFLGIIHHVGNVHVF | 4 | 9 |
| Clavanin-E | FLLGIIHHVGNVHVF | 4 | 9 |
| Clavanin-C | VFLLGIIHHVGNVHVF | 4 | 10 |
| Clavanin-D | FLLGIIHHVGNVHVF | 4 | 9 |
| Aurelin | AACDHICESFKSFCDSGNGVLRKKTCLC | 13 | 8 |
| Dicynthaurin | ILQKAVLDCLKAAGSSLSAIYNKIT | 10 | 12 |
| Halocidin subunit A | WLNLLHHGLNAA | 4 | 7 |
| Halocidin subunit B | LLHHGLNAA | 3 | 5 |
| Tachyplesin II | RWCRVCRGYRKC | 5 | 2 |
| Polyphemusin I | RRWCRVCRGYRKC | 6 | 2 |
| Tachyplesin III | WCRVCRGYRKC | 4 | 2 |
| Polyphemusin II | RRWCRVCKGYRKC | 5 | 2 |
Table 4:The hydrophobic amino acids (red) and hydrophilic ones (blue) distribution through reduced hypothetical peptides sequences.
Specifying the amino acids that had disorder
The amino acids that had disorder in the twenty investigated complete sequences of marine bioeffective peptides were specified from PrDOS as demonstrated in Figure 10. The red color in the sequences signified amino acids with disorder. According to PrDOS, Halocidin subunit B (15 amino acids length) was totally disordered (Figure 10). None of the peptides was
totally ordered, nevertheless Polyphemusin II (18 amino acids) had only one disordered amino acid (Figure 10).
PrDOS could act on all investigated sequences whatever their lengths are (Ishida and Kinoshita, 2007). This algorithm anticipates disorder from sequences of amino acids. The probabilities below 0.5 in the curve showed ordered residues, while those above 0.5 showed disordered ones (Figure 10).
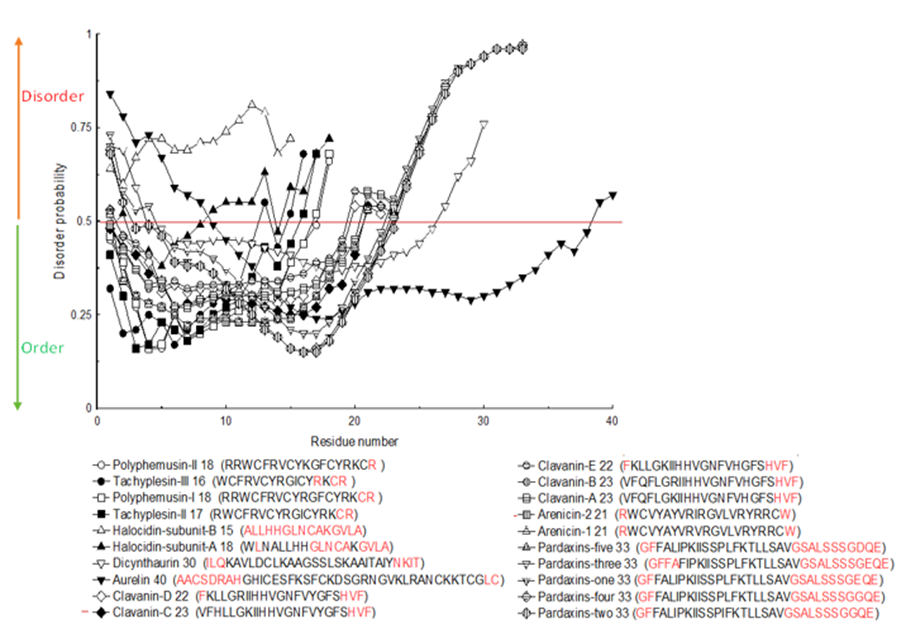
Figure 10: Disorder probability for 20 peptides’ complete sequences obtained from PrDOS server.
The red colored residues in the sequences represent amino acids with intrinsic disorder.
Figure 11 revealed disorder in seven peptides complete sequences (that comprise 30 or more amino acids) obtained from PONDR. The red color in the sequences exemplifies amino acids with disorder. Dicynthaurin is a naturally ordered peptide (via PONDR score). Aurelin had the highest numbers of disordered residues (16 of 40), Pardaxins 1, 3, and 5 had 11 disordered residues, and Pardaxins 2 and 4 had 12 ones (Figure 11). The scores below 0.5 showed ordered residues, while those above 0.5 showed disordered ones (Figure 11).

Figure 11: Intrinsic disorder for seven peptides’ complete sequences (that comprise 30 or more amino acids) obtained from PONDR. The red color in the sequences represents amino acids with disorder.
As can be concluded from Figure 10 and Figure 11, there are differences in the records obtained from both algorithms. These differences originated from the variation in their anticipation method since PONDR depend on matching queries to crystallization data of natural proteins not the sequence itself (Li et al., 1999).
Figure 12 presented flDPnn propensity of disorder in investigated peptides sequences. Records of disorder are vital in designing of de novo or hybrid peptides as they are strictly linked to peptides bioeffectiveness (Dunker et al., 2000, Ward et al., 2004, Shimizu et al., 2007).



Figure 12: flDPnn propensity of disorder in investigated peptides sequences.
This study investigated twenty bioeffective peptides from marine derivation. It focused on comparative exploration for amino acids-% in the different peptides, alignment, phylogeny tree, chemical and physical parameters, hydrophobic/hydrophilic amino acids, cyclic structures based on disulfide bonds, and intrinsic disorder. The study included reducing the original sequences to map the internal similarity that might be hidden in the different peptides. The achieved results can be beneficial in planning de novo or hybrids of these peptides.
N.A.E.-B. and A.A.A.: Conceptualization, Methodology, Data curation and analysis, draft writing. N.A.E.-B. revised and finalized the work. Both authors read and agreed to the manuscript.
The authors declare no conflict of interest.
Clearly Auctoresonline and particularly Psychology and Mental Health Care Journal is dedicated to improving health care services for individuals and populations. The editorial boards' ability to efficiently recognize and share the global importance of health literacy with a variety of stakeholders. Auctoresonline publishing platform can be used to facilitate of optimal client-based services and should be added to health care professionals' repertoire of evidence-based health care resources.

Journal of Clinical Cardiology and Cardiovascular Intervention The submission and review process was adequate. However I think that the publication total value should have been enlightened in early fases. Thank you for all.

Journal of Women Health Care and Issues By the present mail, I want to say thank to you and tour colleagues for facilitating my published article. Specially thank you for the peer review process, support from the editorial office. I appreciate positively the quality of your journal.
Journal of Clinical Research and Reports I would be very delighted to submit my testimonial regarding the reviewer board and the editorial office. The reviewer board were accurate and helpful regarding any modifications for my manuscript. And the editorial office were very helpful and supportive in contacting and monitoring with any update and offering help. It was my pleasure to contribute with your promising Journal and I am looking forward for more collaboration.

We would like to thank the Journal of Thoracic Disease and Cardiothoracic Surgery because of the services they provided us for our articles. The peer-review process was done in a very excellent time manner, and the opinions of the reviewers helped us to improve our manuscript further. The editorial office had an outstanding correspondence with us and guided us in many ways. During a hard time of the pandemic that is affecting every one of us tremendously, the editorial office helped us make everything easier for publishing scientific work. Hope for a more scientific relationship with your Journal.

The peer-review process which consisted high quality queries on the paper. I did answer six reviewers’ questions and comments before the paper was accepted. The support from the editorial office is excellent.

Journal of Neuroscience and Neurological Surgery. I had the experience of publishing a research article recently. The whole process was simple from submission to publication. The reviewers made specific and valuable recommendations and corrections that improved the quality of my publication. I strongly recommend this Journal.

Dr. Katarzyna Byczkowska My testimonial covering: "The peer review process is quick and effective. The support from the editorial office is very professional and friendly. Quality of the Clinical Cardiology and Cardiovascular Interventions is scientific and publishes ground-breaking research on cardiology that is useful for other professionals in the field.

Thank you most sincerely, with regard to the support you have given in relation to the reviewing process and the processing of my article entitled "Large Cell Neuroendocrine Carcinoma of The Prostate Gland: A Review and Update" for publication in your esteemed Journal, Journal of Cancer Research and Cellular Therapeutics". The editorial team has been very supportive.

Testimony of Journal of Clinical Otorhinolaryngology: work with your Reviews has been a educational and constructive experience. The editorial office were very helpful and supportive. It was a pleasure to contribute to your Journal.

Dr. Bernard Terkimbi Utoo, I am happy to publish my scientific work in Journal of Women Health Care and Issues (JWHCI). The manuscript submission was seamless and peer review process was top notch. I was amazed that 4 reviewers worked on the manuscript which made it a highly technical, standard and excellent quality paper. I appreciate the format and consideration for the APC as well as the speed of publication. It is my pleasure to continue with this scientific relationship with the esteem JWHCI.

This is an acknowledgment for peer reviewers, editorial board of Journal of Clinical Research and Reports. They show a lot of consideration for us as publishers for our research article “Evaluation of the different factors associated with side effects of COVID-19 vaccination on medical students, Mutah university, Al-Karak, Jordan”, in a very professional and easy way. This journal is one of outstanding medical journal.
Dear Hao Jiang, to Journal of Nutrition and Food Processing We greatly appreciate the efficient, professional and rapid processing of our paper by your team. If there is anything else we should do, please do not hesitate to let us know. On behalf of my co-authors, we would like to express our great appreciation to editor and reviewers.

As an author who has recently published in the journal "Brain and Neurological Disorders". I am delighted to provide a testimonial on the peer review process, editorial office support, and the overall quality of the journal. The peer review process at Brain and Neurological Disorders is rigorous and meticulous, ensuring that only high-quality, evidence-based research is published. The reviewers are experts in their fields, and their comments and suggestions were constructive and helped improve the quality of my manuscript. The review process was timely and efficient, with clear communication from the editorial office at each stage. The support from the editorial office was exceptional throughout the entire process. The editorial staff was responsive, professional, and always willing to help. They provided valuable guidance on formatting, structure, and ethical considerations, making the submission process seamless. Moreover, they kept me informed about the status of my manuscript and provided timely updates, which made the process less stressful. The journal Brain and Neurological Disorders is of the highest quality, with a strong focus on publishing cutting-edge research in the field of neurology. The articles published in this journal are well-researched, rigorously peer-reviewed, and written by experts in the field. The journal maintains high standards, ensuring that readers are provided with the most up-to-date and reliable information on brain and neurological disorders. In conclusion, I had a wonderful experience publishing in Brain and Neurological Disorders. The peer review process was thorough, the editorial office provided exceptional support, and the journal's quality is second to none. I would highly recommend this journal to any researcher working in the field of neurology and brain disorders.

Dear Agrippa Hilda, Journal of Neuroscience and Neurological Surgery, Editorial Coordinator, I trust this message finds you well. I want to extend my appreciation for considering my article for publication in your esteemed journal. I am pleased to provide a testimonial regarding the peer review process and the support received from your editorial office. The peer review process for my paper was carried out in a highly professional and thorough manner. The feedback and comments provided by the authors were constructive and very useful in improving the quality of the manuscript. This rigorous assessment process undoubtedly contributes to the high standards maintained by your journal.

International Journal of Clinical Case Reports and Reviews. I strongly recommend to consider submitting your work to this high-quality journal. The support and availability of the Editorial staff is outstanding and the review process was both efficient and rigorous.

Thank you very much for publishing my Research Article titled “Comparing Treatment Outcome Of Allergic Rhinitis Patients After Using Fluticasone Nasal Spray And Nasal Douching" in the Journal of Clinical Otorhinolaryngology. As Medical Professionals we are immensely benefited from study of various informative Articles and Papers published in this high quality Journal. I look forward to enriching my knowledge by regular study of the Journal and contribute my future work in the field of ENT through the Journal for use by the medical fraternity. The support from the Editorial office was excellent and very prompt. I also welcome the comments received from the readers of my Research Article.

Dear Erica Kelsey, Editorial Coordinator of Cancer Research and Cellular Therapeutics Our team is very satisfied with the processing of our paper by your journal. That was fast, efficient, rigorous, but without unnecessary complications. We appreciated the very short time between the submission of the paper and its publication on line on your site.

I am very glad to say that the peer review process is very successful and fast and support from the Editorial Office. Therefore, I would like to continue our scientific relationship for a long time. And I especially thank you for your kindly attention towards my article. Have a good day!

"We recently published an article entitled “Influence of beta-Cyclodextrins upon the Degradation of Carbofuran Derivatives under Alkaline Conditions" in the Journal of “Pesticides and Biofertilizers” to show that the cyclodextrins protect the carbamates increasing their half-life time in the presence of basic conditions This will be very helpful to understand carbofuran behaviour in the analytical, agro-environmental and food areas. We greatly appreciated the interaction with the editor and the editorial team; we were particularly well accompanied during the course of the revision process, since all various steps towards publication were short and without delay".

I would like to express my gratitude towards you process of article review and submission. I found this to be very fair and expedient. Your follow up has been excellent. I have many publications in national and international journal and your process has been one of the best so far. Keep up the great work.

We are grateful for this opportunity to provide a glowing recommendation to the Journal of Psychiatry and Psychotherapy. We found that the editorial team were very supportive, helpful, kept us abreast of timelines and over all very professional in nature. The peer review process was rigorous, efficient and constructive that really enhanced our article submission. The experience with this journal remains one of our best ever and we look forward to providing future submissions in the near future.

I am very pleased to serve as EBM of the journal, I hope many years of my experience in stem cells can help the journal from one way or another. As we know, stem cells hold great potential for regenerative medicine, which are mostly used to promote the repair response of diseased, dysfunctional or injured tissue using stem cells or their derivatives. I think Stem Cell Research and Therapeutics International is a great platform to publish and share the understanding towards the biology and translational or clinical application of stem cells.

I would like to give my testimony in the support I have got by the peer review process and to support the editorial office where they were of asset to support young author like me to be encouraged to publish their work in your respected journal and globalize and share knowledge across the globe. I really give my great gratitude to your journal and the peer review including the editorial office.

I am delighted to publish our manuscript entitled "A Perspective on Cocaine Induced Stroke - Its Mechanisms and Management" in the Journal of Neuroscience and Neurological Surgery. The peer review process, support from the editorial office, and quality of the journal are excellent. The manuscripts published are of high quality and of excellent scientific value. I recommend this journal very much to colleagues.

Dr.Tania Muñoz, My experience as researcher and author of a review article in The Journal Clinical Cardiology and Interventions has been very enriching and stimulating. The editorial team is excellent, performs its work with absolute responsibility and delivery. They are proactive, dynamic and receptive to all proposals. Supporting at all times the vast universe of authors who choose them as an option for publication. The team of review specialists, members of the editorial board, are brilliant professionals, with remarkable performance in medical research and scientific methodology. Together they form a frontline team that consolidates the JCCI as a magnificent option for the publication and review of high-level medical articles and broad collective interest. I am honored to be able to share my review article and open to receive all your comments.

“The peer review process of JPMHC is quick and effective. Authors are benefited by good and professional reviewers with huge experience in the field of psychology and mental health. The support from the editorial office is very professional. People to contact to are friendly and happy to help and assist any query authors might have. Quality of the Journal is scientific and publishes ground-breaking research on mental health that is useful for other professionals in the field”.

Dear editorial department: On behalf of our team, I hereby certify the reliability and superiority of the International Journal of Clinical Case Reports and Reviews in the peer review process, editorial support, and journal quality. Firstly, the peer review process of the International Journal of Clinical Case Reports and Reviews is rigorous, fair, transparent, fast, and of high quality. The editorial department invites experts from relevant fields as anonymous reviewers to review all submitted manuscripts. These experts have rich academic backgrounds and experience, and can accurately evaluate the academic quality, originality, and suitability of manuscripts. The editorial department is committed to ensuring the rigor of the peer review process, while also making every effort to ensure a fast review cycle to meet the needs of authors and the academic community. Secondly, the editorial team of the International Journal of Clinical Case Reports and Reviews is composed of a group of senior scholars and professionals with rich experience and professional knowledge in related fields. The editorial department is committed to assisting authors in improving their manuscripts, ensuring their academic accuracy, clarity, and completeness. Editors actively collaborate with authors, providing useful suggestions and feedback to promote the improvement and development of the manuscript. We believe that the support of the editorial department is one of the key factors in ensuring the quality of the journal. Finally, the International Journal of Clinical Case Reports and Reviews is renowned for its high- quality articles and strict academic standards. The editorial department is committed to publishing innovative and academically valuable research results to promote the development and progress of related fields. The International Journal of Clinical Case Reports and Reviews is reasonably priced and ensures excellent service and quality ratio, allowing authors to obtain high-level academic publishing opportunities in an affordable manner. I hereby solemnly declare that the International Journal of Clinical Case Reports and Reviews has a high level of credibility and superiority in terms of peer review process, editorial support, reasonable fees, and journal quality. Sincerely, Rui Tao.
